Previous Projects
Population genetics and phylogeography of brown trout in Austria.
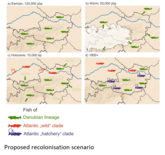
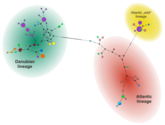
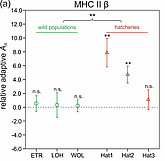
This project aimed to describe the genetic composition of wild and farmed populations of brown trout in Austria. A high-resolution typing protocol was developed, including 17 microsatellite loci and sequencing of >6,000 bp of mitochondrial DNA. Examination of several wild populations led us to formulate the following hypothesis about the postglacial recolonization of brown trout in Austria : First, there was a postglacial expansion of the range of the so-called Danubian genetic lineage into large parts of Austria. In a second wave of range expansions, the Atlantic lineage also migrated into areas of Upper and Lower Austria. During the 20th century, another Atlantic lineage was introduced by extensive stocking by humans. Between the pure Danubian populations we found strong differences and splitting times, which date back to the last interglacial (about 120,000 years ago). While autochthonous wild populations seem to be more influenced by genetic drift, we observed relatively stronger selection pressure on adaptive loci in breeding populations. Because environmental conditions in farms are usually drastically different from those in the wild, this selection pressure could favor genetic variants that are detrimental to fish exposed in a natural environment.
eDNA assesment on the fish fauna in the Ötztaler Ache river using metabarcoding
The fish fauna of the Ötztaler Ache river was surveyed via eDNA metabarcoding. For this purpose, water samples were taken at 28 locations and the environmental DNA contained therein was extracted. This is amplified using universal fish primers and sequenced using illumina technology.
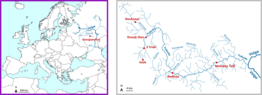
SSqeDNA - Single species targeting via qPCRs in fish monitoring in the headwaters of the Volga river using eDNA
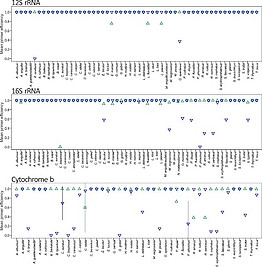
As part of the research project "MABeDNA" - Monitoring Aquatic Biodiversity in the headwaters of the Volga River using eDNA", water samples were taken for eDNA analysis at 11 sample sites in the upper reaches of the Volga River. Total DNA was extracted from these samples and metabarcoding was used to determine the fish fauna present in the respective waters. In this process 23 native but also introduced species of these waters could be successfully detected. Four fish species (sterlet - Acipenser ruthenus, eel - Anguilla anguilla, catfish - Silurus glanis and brook lamprey - Lampetra planeri) could not be detected by metabarcoding in the investigated waters. However, there is other evidence or suitable habitat for these species at several sampling sites. In this project, the metabarcoding results were re-evaluated after expanding the genetic reference database and verified by targeted detection approaches (using qPCR). Four additional fish species were detected in this system and important technical aspects of the metabarcoding approach were elucidated.